What is an operon?
An operon is a set of genes that are regulated together and controlled by a single promoter. This unit of genetic material consists of a promoter, operator, and structural gene(s) that are transcribed into mRNA together. Operons are commonly found in prokaryotic organisms (e.g., bacteria) and some eukaryotes (e.g., Caenorhabditis elegans) [1,2]. The “on” or “off” switch for operon expression is an important adaptation as it allows cells to respond to environmental changes actively and efficiently. The first described operon was the lactose operon in Escherichia coli, also commonly referred to as the lac operon [3].
How does the lac operon work?
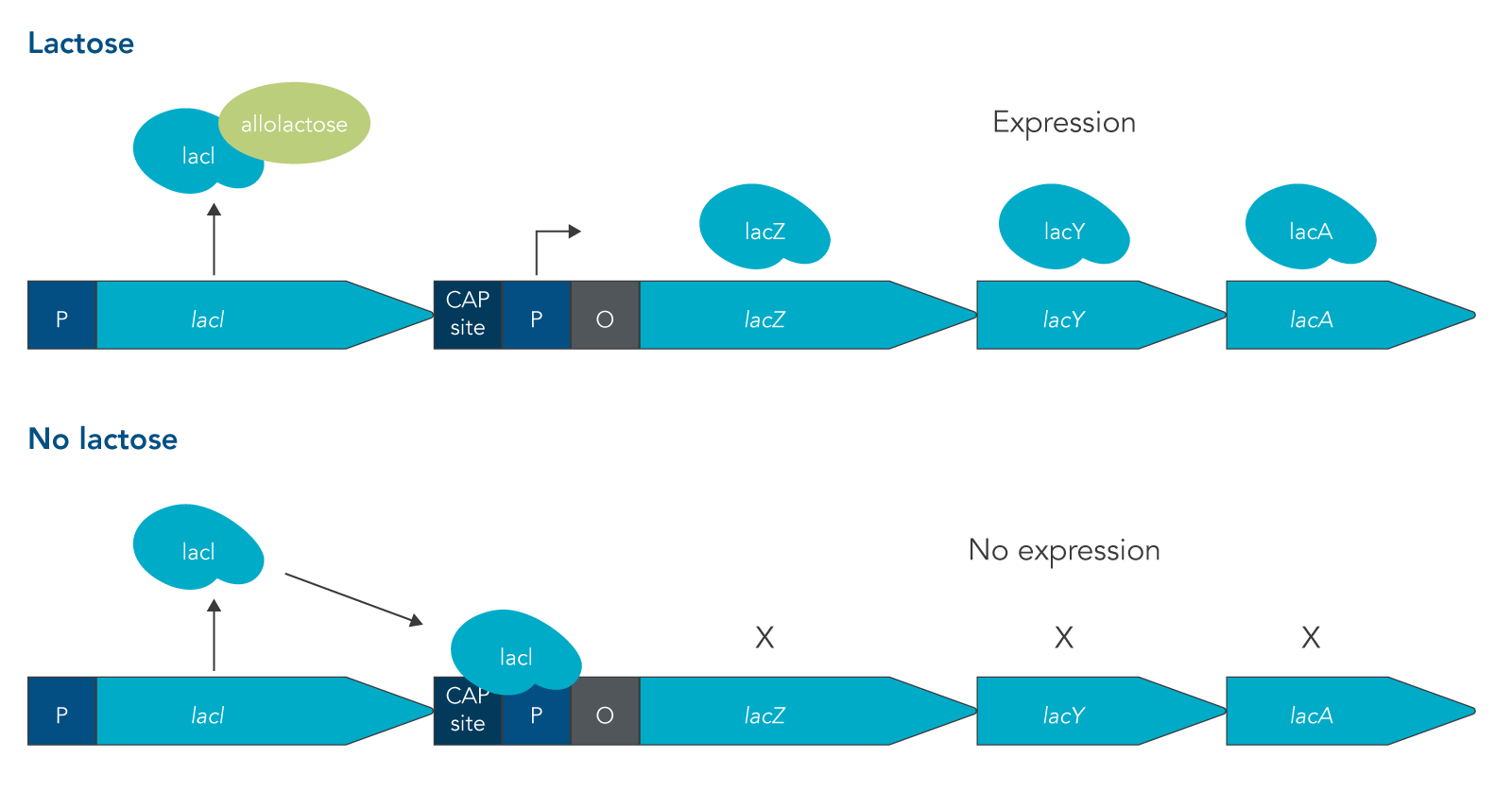
The lac operon is required for the transport and metabolism of lactose. The lacI gene is constitutively expressed, meaning that it is continuously transcribed by cells whether lactose is present or not. In the absence of lactose, the LacI protein binds to the promoter of the lac operon, preventing RNA polymerase (RNAP) from binding. This means that the rest of the lac operon (lacZ, lacY, and lacA) can’t be expressed. Because lactose is not metabolized as easily as glucose, the presence of lactose in an environment is not enough to turn on expression of the lac operon. Instead for the lac operon to be expressed, lactose must be present and glucose concentrations must be low or absent.
Under these conditions, two things happen that allow for the structural genes of the lac operon to be expressed:
- LacI binds with a disaccharide that is structurally similar to lactose called allolactose. When this binding occurs, LacI is released from the promoter sequence.
- The catabolite activator protein (CAP) is expressed and forms a complex with cyclic adenosine monophosphate (cAMP). cAMP is a molecule that is present in high concentrations when glucose levels are low.
This double requirement prevents the lac operon from being expressed when an easier to metabolize sugar like glucose is present, saving energy for cells to express other more important genes [4]. The structural genes that are expressed in the lac operon are lacZ, lacY, and lacA (Figure 1). The lacZ gene produces the enzyme beta-galactosidase, lacY produces the beta-galactoside permease protein, and lacA encodes for the beta-galactoside transacetylase enzyme. LacZ breaks down lactose into glucose and galactose; while LacY is responsible for bringing lactose into the cell; and LacA transfers an acetyl group from coenzyme A to a hydroxyl group of a galactoside [4].
It is worth noting that the lac operon is also expressed when analogues of lactose like isopropyl β‑D-1-thiogalactopyranoside (IPTG) are present. This characteristic has made the lac operon an important tool for mutant selection.
Lac operon and blue white screening
Blue white screening is an approach that takes advantage of the lac operon and allows researchers to easily identify viable colonies for downstream cloning steps. The blue white screening technique uses competent E.coli cells that are missing a functional lacZ gene as well as a cloning vector that contains a modified lacZ gene and an antibiotic resistance gene. The lacZ gene within the cloning vector contains a multiple cloning site (MCS) which allows for a fragment of DNA to be inserted into the middle of the gene.
After the transformation step in a cloning protocol, bacterial cells are grown on agar plates that have an antibiotic, IPTG, and a molecule called 5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside (x-gal). The antibiotic helps select against the cells that survived transformation but did not accept the plasmid. IPTG functions as an inducer of the lac operon, and x‑gal helps researchers determine whether the cells contain recombinant DNA.
Bacterial colonies with an empty plasmid (i.e., plasmids without a DNA insert) turn blue. This occurs because of a process called alpha-complementation in which the lacZ gene in the cloning vector complements the lacZ deficient E.coli cells. The gene expression of lacZ is induced by IPTG and the resulting LacZ enzyme can breakdown x-gal into galactose. Importantly, when x-gal is catabolized the byproduct, 5-bromo-4-chloro-indoxyl, dimerizes producing an insoluble blue pigment.
However, colonies made up of cells that accept a plasmid with the DNA insertion remain white, because the DNA that was inserted into the cloning vector at the MCS disrupts the lacZ gene making it impossible for the cell to create a functional enzyme to metabolize x-gal.
In other words, blue white screening enables researchers to quickly assess a large number colonies using the naked eye. Bacterial colonies that appear white on agar plates can be further analyzed using colony PCR and/or sequencing to confirm that the inserted plasmid was correctly copied and assembled.


