DNA cloning
Overview
DNA cloning—often referred to as molecular cloning—is a broad term encompassing various experimental techniques designed to isolate a specific DNA fragment and propagate it within a host organism, ultimately producing numerous identical copies of recombinant DNA.
DNA cloning was first described in the 1970s and has since evolved to become the cornerstone of biotechnology.

What is DNA Cloning?
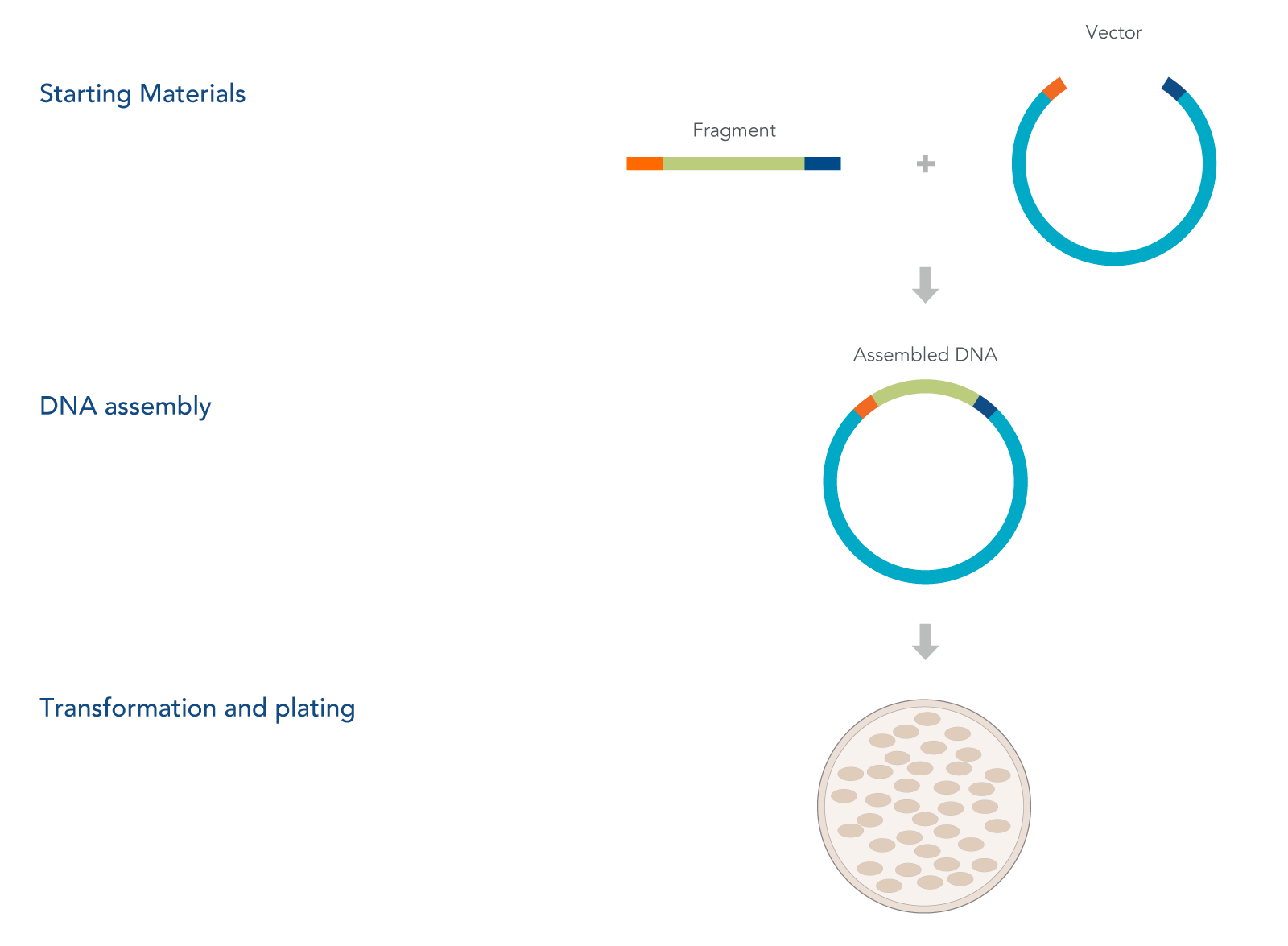
DNA cloning involves a series of steps, beginning with the generation of recombinant DNA and vector constructs, followed by their introduction into host cells, and concluding with verification of the desired recombinant product. This process typically starts by joining DNA fragments and inserting them into a suitable vector—such as plasmids, attenuated viruses, bacteriophages, or larger constructs like artificial chromosomes. The recombinant DNA construct is then delivered into host cells, where the cellular machinery replicates, and in some cases, expresses the introduced sequence.
This recombinant DNA technology is the foundation for a range of synthetic biology products including vaccines, biologics, enzymes, improved crop varieties, microbial strains for bioremediation, research models, biodegradable materials, and biofuels, among others.
What is a DNA cloning workflow?
The basic cloning workflow involves these five steps:
- Designing and isolating the DNA sequence of interest (insert)
- Selecting the vector into which the insert will be introduced
- Using a cloning method to introduce the insert into the vector
- Transforming the cloned construct into host cells
- Screening and selecting host cell clones containing the correct recombinant DNA construct
Types of cloning methods
Recombinant DNA cloning encompasses multiple methods for inserting a desired DNA fragment into a chosen vector, each offering distinct advantages and limitations. Selecting the appropriate technique—whether blunt-end, TA, restriction enzyme-based, Golden Gate, seamless assembly (e.g., Gibson Assembly®), or Gateway®—is a critical aspect of experimental design, as it can significantly influence cloning efficiency and downstream applications.
- Blunt-end cloning: Involves inserting DNA fragments with blunt termini into a linearized vector, using DNA ligase to covalently join the 3′-hydroxyl and 5′-phosphate ends.
- TA cloning: Leverages Taq polymerase to generate single-base overhangs, enabling the insertion of DNA fragments into a vector through complementary A–T pairing.
- Restriction enzyme cloning: Uses sticky ends generated by endonuclease digestion to facilitate the insertion of DNA fragments into a compatible vector through complementary base pairing.
- Golden Gate cloning: Uses type IIS restriction enzymes to generate complementary sticky ends, enabling precise and directional assembly of DNA fragments into a vector.
- Seamless cloning: Also known as Gibson Assembly®, enables the insertion of DNA fragments into a vector by exploiting complementary overlaps at the terminal ends of both the fragments and the vector.
- Gateway® cloning: A recombination-based technique that enables efficient transfer and shuffling of DNA fragments between compatible vectors.
How are synthetic DNA fragments used for cloning and assembly?
Synthetic, double-stranded DNA fragments provide a convenient alternative to PCR products for cloning and gene assembly applications. These fragments are highly flexible and customizable, allowing researchers to design and validate sequences in silico and tailor them to specific requirements of their chosen cloning method. By eliminating the need to amplify or assemble templates already available in the lab, synthetic DNA offers a faster, more streamlined approach to building constructs.
IDT synthetic DNA fragment solutions for cloning methods
We offer three types of DNA fragments that are customizable and compatible with all cloning applications:
DNA Cloning Guide
This guide discusses common DNA cloning techniques, important sequence design considerations, and tips for troubleshooting that may arise when performing a cloning experiment.