Targeted RNA sequencing
Targeted RNA sequencing with exome, custom, or predesigned hyb capture panels has high mapping rates, high on-target percentage, and low duplication rates. Targeted RNA sequencing decreases intronic and intergenic reads to focus your data on exonic sequences.
xGen™ NGS—made for targeted RNA sequencing.
Overview
- Fast and reliable—get RNA-seq libraries ready for hybridization capture in 3.5 hours
- Provides comprehensive data—for mapping, on-target, and transcript coverage
- Compatible with a range of input types and quantities—prepare RNA-seq libraries from low-input and/or low-quality RNA samples using the xGen Broad‑Range RNA Library Prep Kit
- Consistent libraries—minimal adapter dimers, so adapter titration is not required
- Variety of indexing options available—for a total of 1536 Unique Dual Index (UDI) primer pairs
- Flexible and customizable—choose between different hybridization panels depending on your research needs
- Automation-friendly RNA-seq workflow
What is targeted RNA sequencing?
Targeted RNA sequencing (RNA-seq) is a cost-effective tool for deep sequencing of specified regions of interest within the transcriptome. This approach provides deep sequencing of targeted regions while omitting the undesired regions that often result in a high number of sequencing reads not relevant to the research study.
Reliable, time- and cost-effective approach to targeted RNA-seq workflow
RNA-seq provides the researcher with data about the RNA molecules in the sample at a specific time point—a snapshot in time. In short, the workflow consists of RNA extraction, rRNA depletion or mRNA enrichment, cDNA conversion, library preparation, and finally, sequencing. Sometimes identifying all RNAs in a sample is beyond the scope of the experiment, and a strategy to focus the library to regions or targets of interest is preferred. In targeted RNA-seq, the traditional workflow can be modified to consist of RNA extraction, cDNA conversion, and library preparation, followed by hybridization capture with either a custom or predesigned hyb cap panel.
For library prep, IDT offers two different solutions that include cDNA conversion.
- For regular RNA samples, the IDT xGen RNA Library Prep Kit offers a fast NGS transcriptomic research workflow. RNA-seq libraries can be produced using tissue samples, blood, or high-quality RNA samples with an RNA Integrity Number (RIN) of 7–10. This RNA-seq workflow also utilizes Adaptase™ technology to produce libraries following first-strand cDNA synthesis (see the xGen RNA Library workflow). This workflow can be easily combined with xGen UDI Primer Plates for indexing.
- The xGen Broad‑Range RNA Library Prep Kit supports a wide input range, including low-quality formalin-fixed paraffin-embedded (FFPE) RNA samples. This stranded RNA-seq workflow for low-quality and/or low-input RNA samples utilizes Adaptase technology to produce libraries following first-strand cDNA synthesis (refer to xGen Broad-Range RNA Library workflow). Samples are recommended to have a RIN >2 or DV200 >30 (DV200 is a quality metric that represents the percentage of RNA fragments longer than 200 nucleotides). This workflow can also be easily combined with xGen UDI Primer Plates for indexing.
After library prep, hybridization capture with IDT xGen Hyb Probes, which are individually synthesized, 5’-biotinylated oligos, enriches for fragments corresponding to the targets of interest. For researchers interested in the mRNA subset expressed from exon regions of the genome, you can pair the xGen Broad‑Range RNA Library Prep Kit or the xGen RNA Library Prep Kit with the xGen Exome Hyb Panel v2 for more efficient expression profiling, yielding a higher percentage of coding bases and a lower percentage of intronic bases than whole transcriptome data (see Figure 1B).
For researchers interested in a more focused target space, IDT offers a variety of other xGen Predesigned Hybridization Capture Panels, and if these are not exactly what you want, an IDT xGen Custom Hyb Panel can be designed to target your specific genetic regions of interest. IDT also offers the xGen Hybridization and Wash v2 Kit that includes buffers, Cot DNA, our xGen 2X HiFi PCR Mix, and streptavidin-coated magnetic beads (note, the beads and reagents cannot be ordered separately).
In order to prevent non-specific pull-down of fragments during the hybridization capture reaction, IDT has developed xGen Universal Blockers that inhibit adapter:adapter annealing that can be one of the causes of non-specific interactions.
Extraction
Library Prep
Library prep kits
Indexing primers
Hybridization Capture
xGen Hybridization Panels
xGen Hybridization Capture Core Reagents
Sequencing & analysis
Method data
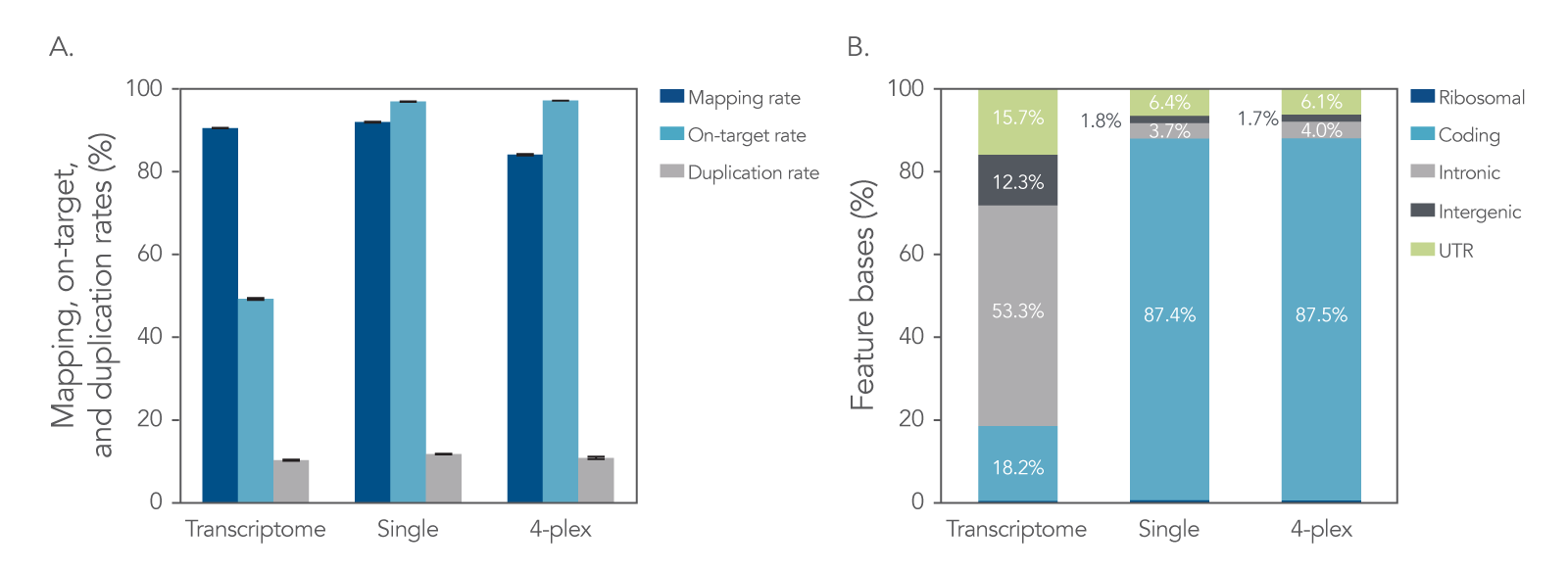
Libraries were prepared with the xGen Broad-Range RNA Library Prep Kit using a degraded FFPE RNA sample. The libraries were used as a single-plex or in a 4-plex hybridization capture with the predesigned xGen Exome Hyb Panel v2. An efficient library prep and hybridization capture workflow results in high mapping rates, high on-target percentages, and low duplication rates. Captured mRNA libraries result in an increased number of exonic sequencing reads due to the decreased amount of intergenic and intronic reads when compared to transcriptome libraries. This translates to improved sequencing cost efficiency by reducing the number of unusable reads. All libraries had a removal of >99% of rRNA bases, indicating that ribodepletion is unnecessary for hybridization captured libraries (Figure 1).
Figure 1. Target enrichment metrics. The xGen Broad-Range RNA Library Prep Kit was used to generate libraries from extracted FFPE RNA. For the transcriptome condition, libraries were prepared from 10 ng of rRNA-depleted RNA and directly sequenced (n = 3). For the hybridization capture conditions (single-plex and 4-plex), libraries were prepared from 50 ng of total RNA (n = 7). Subsequently, three libraries underwent hybridization capture individually (as single-plex captures) and four libraries were pooled together for a 4-plex capture using the xGen Exome Hyb Panel v2, after which the captured libraries were sequenced. Sequencing was performed using a NextSeq® (Illumina) 300 cycle kit and subsampled to 40 M reads per sample. (A) The data show comparable mapping and duplication rates, as well as higher exonic reads and lower intronic reads for hybridization captured samples. Error bars represent the mean ± standard deviation of replicates. (B) The feature bases chart shows that hybridization capture results in a higher percentage of coding bases sequenced, and fewer intronic bases than transcriptome libraries (values within bars represent means of the replicates). Post-capture libraries from these figures were amplified using KAPA 2x HiFi PCR Mix, as these data were generated prior to release of xGen 2x HiFi PCR Mix.
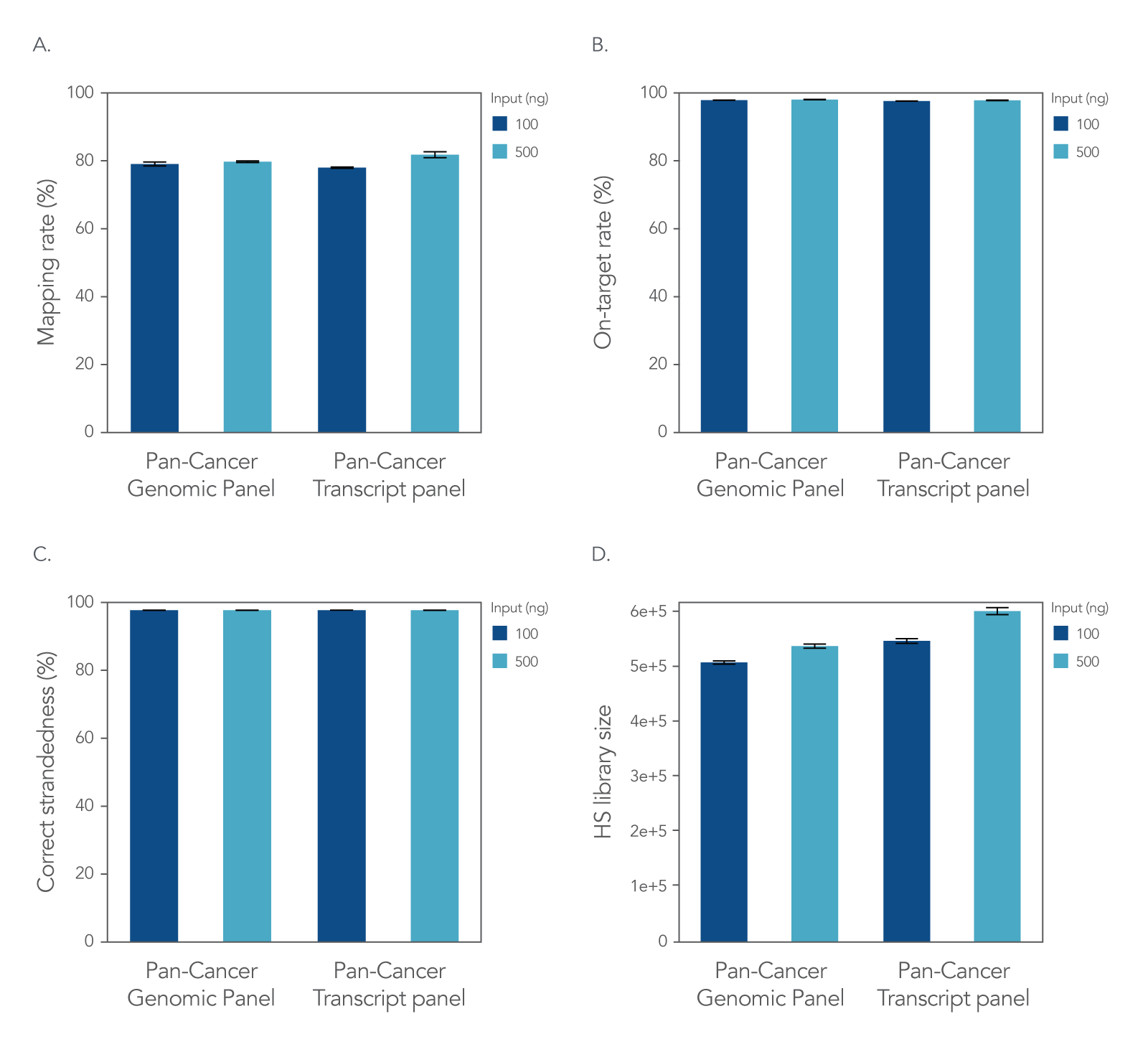
xGen RNA Library Prep Kit with xGen Custom Hyb Panels
Figure 2. xGen Custom Hyb Panels provide high quality sequencing results according to NGS performance metrics. The xGen RNA Library Prep Kit was used to generate sequencing libraries with 100 ng or 500 ng of total UHR RNA followed by hybridization capture using one of two xGen Custom Hyb Capture Panels (Pan-Cancer Genomic Panel for RNA or DNA applications and Pan-Cancer Transcript Panel for RNA applications), and then subsampled to 2 M reads per sample. Sequences were processed and aligned using STAR [1] and the hg38 reference genome [2]. This workflow resulted in (A) a mapping rate of >78%, (B) on-target rate of >98%, and (C) correct strandedness of >99.8%. (D) Sequencing data demonstrate that the number of unique molecules [hybrid-selection (HS) library size] increases as expected with higher RNA input into library prep (Picard) [3]. Post-capture libraries from these figures were amplified using KAPA 2x HiFi PCR Mix, as these data were generated prior to release of xGen 2X HiFi PCR Mix. Error bars in all four panels represent the mean ± standard deviation of replicates.
Ordering
Use our xGen Hyb Panel Design Tool to create your custom hybridization capture panel.
Resources
References
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21.
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006.
- “Picard Toolkit.” Broad Institute, GitHub repository: Broad Institute; 2019. https://broadinstitute.github.io/picard/