IDT offers a powerful analytical tool for determining the properties of oligo sequences—the OligoAnalyzer Tool. This tool calculates the physical characteristics of any oligo sequence, including length, GC content, melting temperature (Tm), molecular weight, extinction coefficient, and optical density. Access the OligoAnalyzer Tool from the “Tools” menu on top of any page on the IDT website (Figure 1).
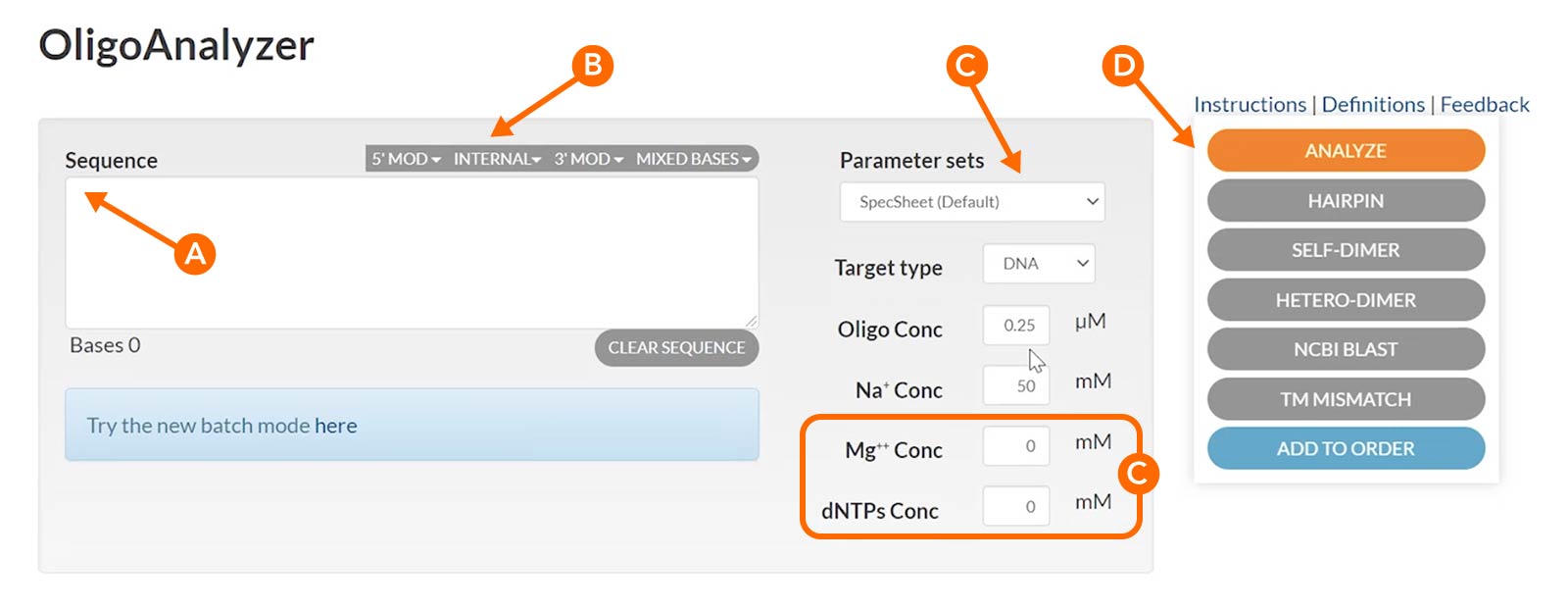
Enter your sequence into the “Sequence” box in the 5’ to 3’ orientation (Figure 2, arrow A). The OligoAnalyzer Tool accommodates DNA, RNA, mixed bases, and a variety of modifications, which can be selected from the menus under the Sequence box (Figure 2B). To get the correct Tm, it is important to enter the Mg++ and dNTP concentrations you will use in your experiment (Figure 2, arrow and box C). Note that the Tm reported on the specification sheet shipped with IDT oligos uses default settings of 0 nM Mg++ and 0 mM dNTPs, so will undoubtedly differ from the Tm calculated here. When you press “Analyze” (Figure 2, arrow D) the tool will produce the complementary sequence, GC content, Tm, molecular weight, and extinction coefficient for you. Additionally, you can run Self-Dimer and Hairpin analyses of your sequence from the Sequence entry screen (below Figure 2, arrow D).
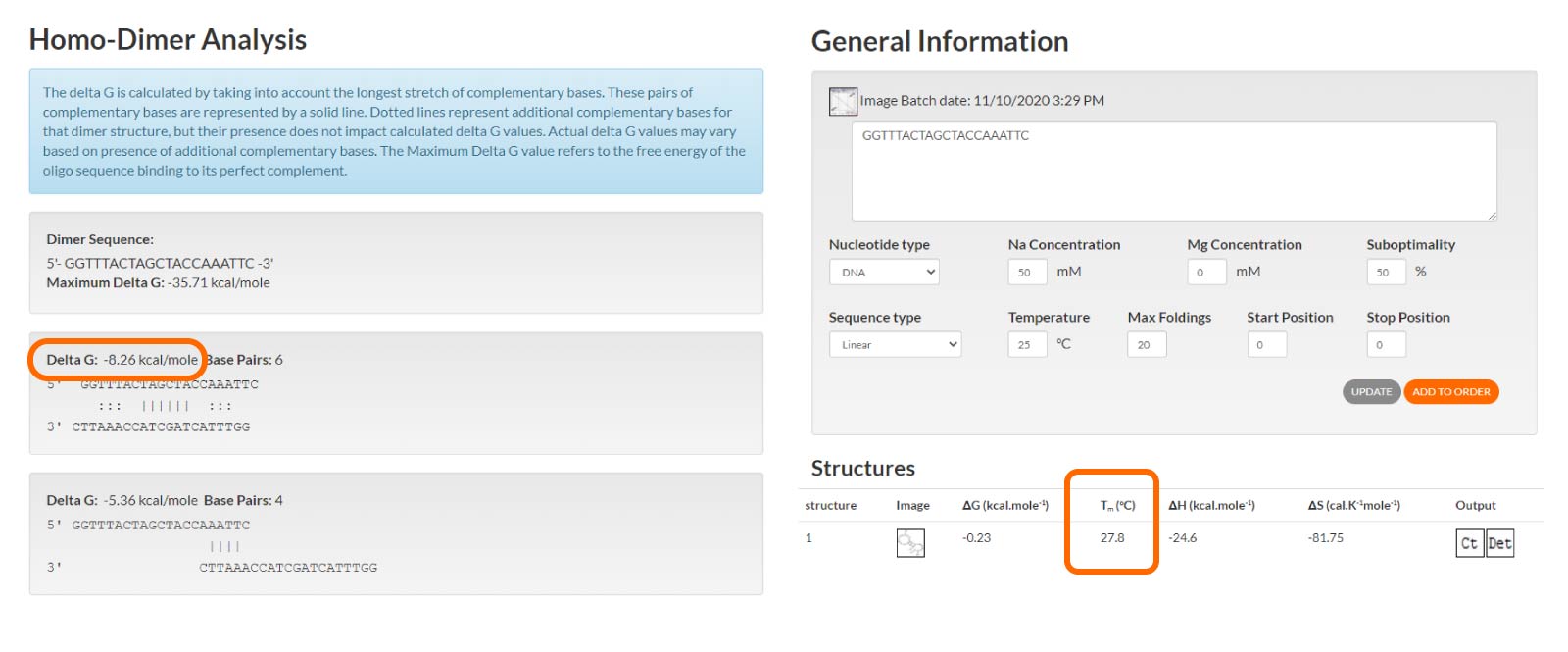
The Gibbs free energy (ΔG) value gives an indication of the strength of the secondary structure. IDT recommends the ΔG value be more than –9 for self-dimers and hetero-dimers. For hairpins, the Tm should be lower than the temperature at which the oligo will be used (Figure 3).
If you need help with the program, click the “Instructions” link for step-by-step guidance on how to use the OligoAnalyzer Tool. The “Definitions” link provides an explanation of terms and equations the program uses to determine Tm and the extinction coefficient (Figure 4).
Go to our login page to learn more. And, of course, you can always contact us.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID: RUO23-1917_001

.jpg?sfvrsn=768cfc07_4)
