rhAmpSeq™ Design Tool
Deep, targeted amplicon sequencing with highly multiplexed panels for sequencing on Illumina® platforms.
- Generate sequencing panels for a wide range of plant and animal species
- Minimize primer dimers and maximize multiplexing capability
- Efficiently analyze on- and off-target CRISPR edit sites
- Multiplex up to 5000-plex in a single pool
* Not all genomes supported by the rhAmpSeq Design Tool are supported by the rhAmpSeq CRISPR Analysis Tool. See below for details.

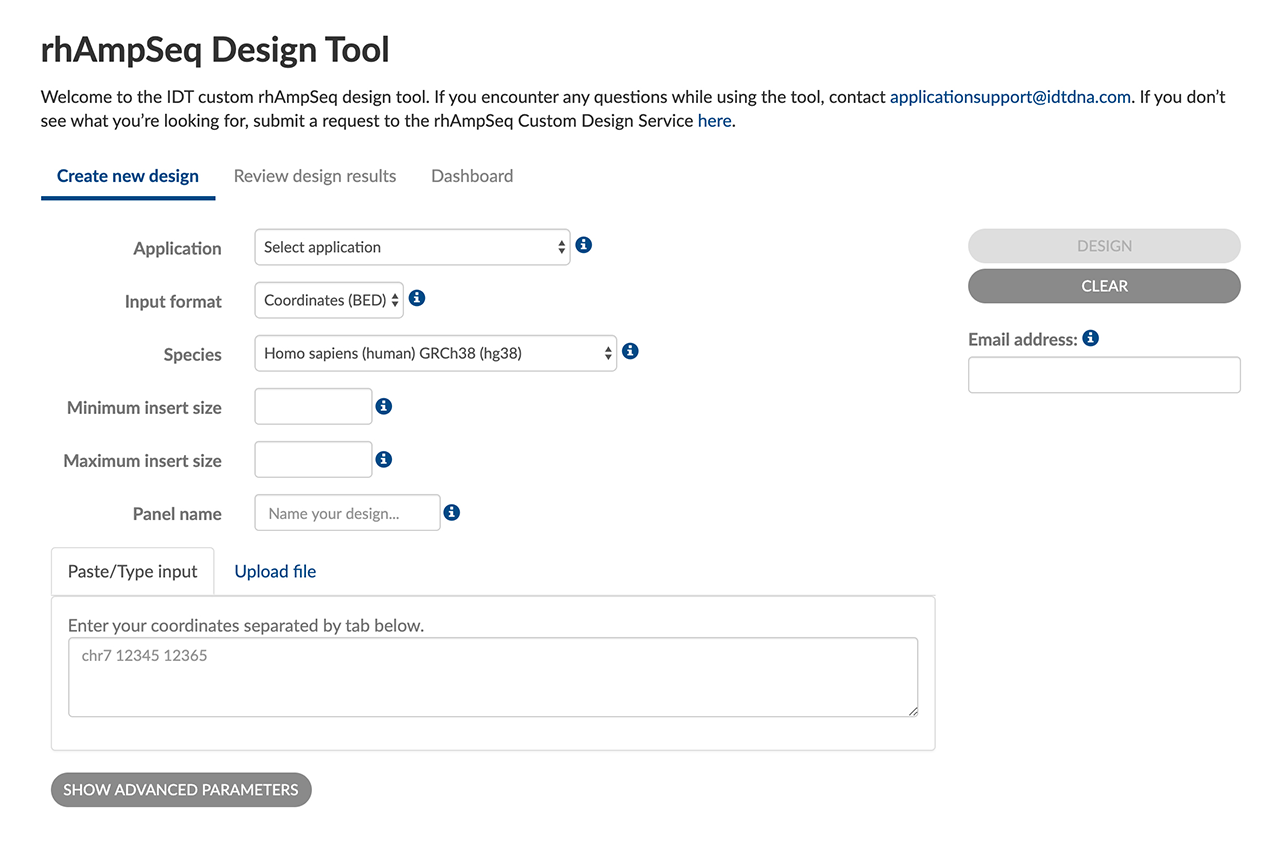
Research-friendly UX
Flexible tool input formats and advanced parameters help you create custom, targeted sequencing panels optimized for your targets:
- Choose BED or FASTA inputs
- Optimize for SNP hotspot or CRISPR indel screening
- Name & save designs for easy reuse and reordering
Generating NGS-ready amplicon libraries just got easier
Design for hotspot target SNPs, indels, and more
Obtain panel designs within 1 hour for most applications*
Accelerate discovery with quicker, easier ordering
Flexible analysis
Below are the available genomes supported by the rhAmpSeq analysis tool. While the design tool can support a larger set of genomes this should be considered if you plan to use the analysis tool
| Genomes supported by the rhAmpSeq Analysis Tool |
|---|
| Caenorhabditis elegans – round worm (WBcel235) (IDT) |
| Danio rerio – zebrafish (GRCz11) (IDT) |
| Homo sapiens – human (GRCh37) (IDT) |
| Homo sapiens – human (GRCh38) (IDT) |
| Macaca fascicularis – crab-eating macaque (macFas5) (IDT) |
| Macaca mulatta – rhesus monkey (rheMac8) (IDT) |
| Mus musculus – mouse (GRCm38) (IDT) |
| Rattus norvegicus – rat (Rnor6) (IDT) |
Tailored to your research
Custom rhAmpSeq Panels have been designed and tested in a wide range of animal and plant species and diverse targeted NGS applications, including:
- Marker-assisted selection and genomic selection in agricultural genetics and breeding
- Confirmation of on- and off‑target CRISPR gene editing experiments
- Hotspot panels for oncology, rare and inherited diseases, and other disease research
Resources
User guides and protocols
Sample data
After years of using traditional amplicon sequencing in grapevine, we found that rhAmpSeq’s specificity and evenness of read depth across sites and samples enabled us to multiplex to much higher levels. IDT provided agricultural pricing that enables us to apply 2000 core genome rhAmpSeq haplotype markers to genetic mapping and marker-assisted selection for grape scientists worldwide. ”
Professor of Grapevine Genetics and Family Genealogist
Cornell University
