The Alt-R™ CRISPR-Cas9 genome editing system
Overview
The combination of Cas9 plus guide RNA forms a ribonucleoprotein (RNP). In nature, this RNP cleaves viral DNA as part of a bacterial immune system. In CRISPR genome editing, scientists have repurposed Cas9 RNP to cut targeted sites in any genome. Alt-R Cas9 enzymes from IDT provide high-potency precise genome editing.

CRISPR-Cas9 genome editing
CRISPR genome editing provides a fast, efficient method of altering the genome of living cells and organisms. The term CRISPR stands for "clustered regularly interspaced short palindromic repeats," which are DNA sequences in bacteria. These sequences function as part of a bacterial defense system, along with CRISPR-associated (Cas) nucleases. The best known and most widely used Cas nuclease is Cas9. This endonuclease creates a double-strand break in DNA.
The CRISPR basics handbook
Everything you need to know about CRISPR, from A to Z, from theory to practice, for beginners as well as advanced users.
How CRISPR-Cas9 works in bacteria and in genome editing
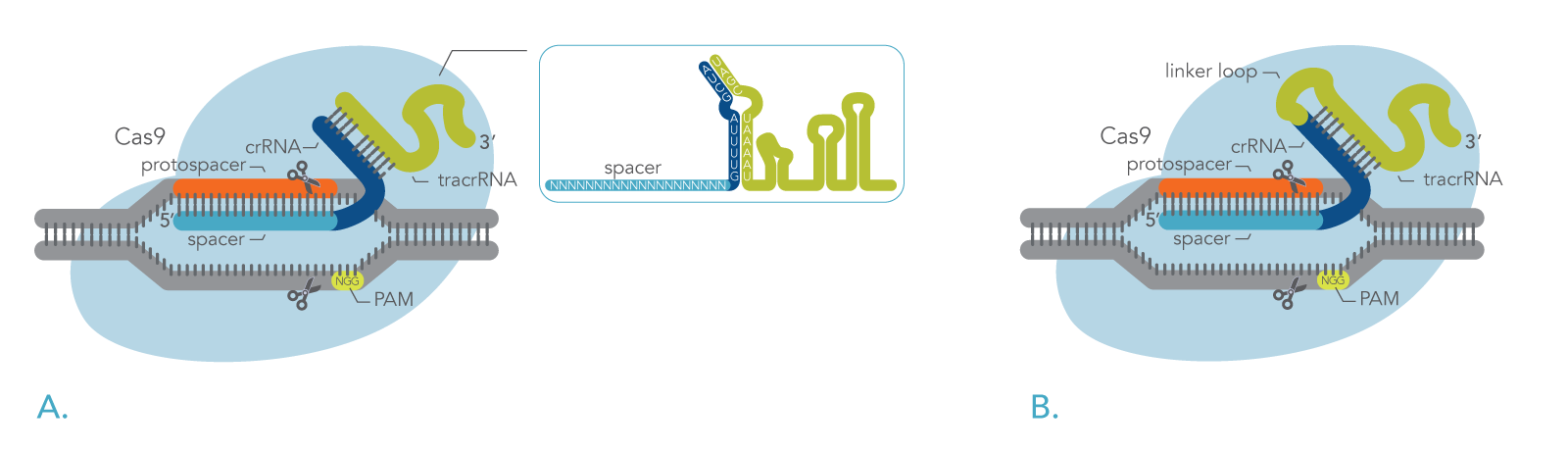
Cas9 is guided to the target site in any DNA (viral DNA, mammalian genomic DNA, etc.) by short, complementary RNA sequences called CRISPR RNAs (crRNAs). The target site, which is an approximately 20-nucleotide DNA sequence, is known as a protospacer. This site is adjacent to a "protospacer-adjacent motif" (PAM), which has the sequence 5′-NGG-3′, where N is any base. The Cas9 enzyme makes a blunt double-strand break (DSB) within the target site, three bases away from the PAM (Figure 1). For CRISPR-Cas9 endonuclease to have activity, the crRNA must be combined with a "trans-activating crRNA" (tracrRNA) to activate the endonuclease and create a functional editing ribonucleoprotein (RNP) complex (Figure 1A).
In bacteria, the RNP recognizes the DNA sequences of invading viruses. The RNP complex cleaves the viral DNA and destroys the virus, protecting the bacteria. This endonuclease activity works the same way in different experimental organisms and cell lines in the lab, so Cas9 can be used for genome editing. The only requirement is to design the crRNA sequence to target a desired genomic site instead of a piece of viral DNA.
Figure 1. DNA cleavage by the CRISPR-Cas9 system. (A) Cas9 with two-part guide RNA. The guide RNA forms a complex with Cas9 endonuclease to guide targeted cleavage of genomic DNA. The cleavage site is specified by the spacer element of the crRNA. The crRNA spacer element recognizes 19 or 20 nt on the strand opposite from the NGG PAM site. The PAM site must be present immediately downstream of the protospacer for cleavage to occur. (B) Cas9 with single-guide RNA. The single-guide RNA (sgRNA) approach uses a fused crRNA and tracrRNA, but otherwise the entire Cas9 system works like the two-part guide RNA system shown in panel A.
As mentioned above, the bacterial Cas9 system uses crRNA and tracrRNA together to form the guide RNA. This is one frequent way in which scientists use Cas9 in genome editing. However, as shown in Figure 1B, in an alternative method also used for genome editing, the crRNA and tracrRNA are synthesized as a single fused RNA oligonucleotide called a single guide RNA (sgRNA). This usually works just as well as a two-part guide RNA system.
IDT offers a couple major options for your Cas9 genome editing work. If you want wild-type Cas9, we offer it as a recombinant protein fused with a proprietary nuclear localization sequence (NLS). We have found that the added NLS improves genome editing. This version of Cas9, known as Alt-R Cas9 Nuclease V3, is an excellent choice for most genome editing experiments. If you need a Cas9 with reduced off-target activity that still has the potency (efficiency) of wild-type Cas9, we additionally offer Alt-R HiFi Cas9 Nuclease, which works well when delivered as an RNP, has very high efficiency, and causes almost no off-target effects.
In addition to Alt-R Cas9 Nuclease V3 and Alt-R HiFi Cas9 Nuclease, IDT also offers other variants of Cas9 (e.g., GFP/RFP variants, nickases, and dead Cas9) that are useful in other kinds of CRISPR experiments.
Positive and negative control gRNA for your Cas9 editing experiments are available in convenient kit format from IDT.
Products for CRISPR-Cas9 genome editing
Alt-R™ CRISPR-Cas9 System
Efficient CRISPR reagents based on the commonly used Streptococcus pyogenes Cas9 system for lipofection or electroporation experiments. Protospacer adjacent motif (PAM) = NGG.
Alt-R™ CRISPR nucleases
Recombinant, high-purity Cas9 and Cas12a (Cpf1) endonucleases for genome editing experiments.
Cas9 guide RNA design tool
Select from our predesigned gRNAs targeting human, mouse, rat, zebrafish, or nematode genes. For other species, use our proprietary algorithms to design custom gRNAs. For protospacer designs of your own or from publications, use our design checker tool to assess targeting potential before ordering gRNAs that are synthesized using Alt-R gRNA modifications.
Cas9 HDR donor template and gRNA design
Provide basic information about your target site, then use the HDR tool to design and visualize your desired edit within the sequence. The HDR Design Tool will provide the recommended gRNA(s) and HDR donor template for your specifications.
Explore resources
RUO22-1417_001