Quick facts:
Availability: DNA and RNA
Location: 5′, internal, and 3′
Synthesis scale: 100 nmol to large scale
Purification options: Standard desalt or HPLC
IDT ordering symbol: /5Super-dG/, /iSuper-dG/, /3Super-dG/
Need an oligo with a poly G run?
When trying to order an oligo with several sequential guanine residues, you will often receive a warning to the effect:
“…Runs of 4 or more G bases can result in a cruciform structure, also known as a guanine tetraplex that may interfere with downstream applications. In addition, oligo sequences containing runs of 6 or more G bases can be difficult to manufacture…”
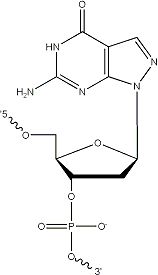
Super G (8-aza-7-deaza dG) bases (Figure 1) are pyrazolo[3,4-d]pyrimidine analogs of guanine that eliminate naturally occurring, non-Watson-and-Crick secondary structures associated with guanine-rich sequences [1].
Including a Super G modified base in place of one of a series of G residues in an oligo sequence:
- Eliminates secondary structures—Poly G runs can form tetraplexes, or cruciform structures, that can cause difficulty with oligo synthesis and use of these oligos in downstream applications [2]. Super G bases prevent these structures from forming [1].
- Increases oligo yields and purity—Substitution of a single G in a 4 G base sequence with a Super G base will improve oligonucleotide synthesis.
- Maintains compatibility with PCR—Primers containing this modified base, even when positioned at the 3′ end, are extended normally by polymerases, including Taq polymerase.
- Improves qPCR signal when adjacent to a fluorophore—Super G bases do not quench fluorophore dyes as can a guanine base adjacent to 5' 6-FAM [3]. In fact, you can observe noticeable improvement in qPCR signal when the G next to a fluorophore is substituted with a Super G base.
- Increases duplex stability and mismatch discrimination—In their paper, Kutyavin IV, Lokhov SG, et al. (2002) [1] investigated the increased Tm Super G base substitutions confer on DNA duplexes when at different positions within a sequence. They also describe improved discrimination of G/A, G/G, and G/T mismatches in Watson–Crick hybrids.
Add Super G bases to your oligo design
To order, enter your sequence with the IDT ordering symbol (shown above) in the desired location within your sequence. Oligos containing Super G bases can be ordered with HPLC purification or as standard desalt, providing that other modifications in the sequence do not require HPLC.
Learn about other modifications to facilitate your research
See what other modifications are available from IDT. You can find a list of standard modifications we make to oligonucleotides on the Modifications page of our online catalog. And if you don’t find what you are looking for, just send us a request. We often synthesize special, non-catalog requests. Contact us at noncat@idtdna.com.